Page 10 - NBAGR NEWSLETTER VOL 16 No 1 (OCT 2019 - JUNE 2020)
P. 10
The risk to the population in a block was categorised based on 1) rate of decline per year and, 2) the probable extinction in the
years. -
No risk : any decline and/or positive growth
Low risk (+) : decline rate is >1.25% and/or annually extinction within 100 years
Moderate risk (++) : decline rate is >2.5% annually and/or extinction within 80 years
High risk (+++) : decline rate is >5.0% annually and/or extinction within 60 years
Very high risk (++++) : decline rate is >10.0% annually and/or extinction within 40 years
Extremely high risk (+++++) : decline rate is >20.0% annually and/or extinction within 20 years
Table-Donkey populations in different blocks of Leh district
during 2012 and 2019 census
All the blocks reported the population
Name of the 2012 2018 Decline Probable Risk status*
Blocks rate (per extinction in of Ladakhi donkeys in the field areas.
year) year after 2020 However, except in Turtuk, the population
Saspol 142 16 36.4 6 +++++ of donkeys have declined in all blocks, with
an annual rate of decline ranging from
Nimoo 281 160 9.4 52 +++ 1.3% in Chumathang to extremely high
Khaltsi 1807 771 14.2 45 ++++ 30.5% in Thiksey block. Khaltsi block, which
Chouchod 339 101 20.2 21 +++++ was carrying highest donkey population
Thiksey 87 14 30.5 7 +++++ was worst affected showing a decline
of more than half. Population in Saspol,
Kharu 129 69 10.5 39 ++++ Chouchod, Thiksey, Panamic, Leh, and
Diskit 757 507 6.7 91 +++ Drubuk would be almost zero in coming
Panamic 225 63 21.2 18 +++++ 20 years. Overall annual decline rate of
Turtuk 531 584 Nil Ladakhi donkey in Leh district is very high
(11.5%), and along with small population
Leh 473 125 22.2 20 +++++ base, the population is in very high risk
Chumathang 80 74 1.3 329 + of extinction, within “Critical” category as
Nyoma 230 117 11.3 40 ++++ per the guidelines laid by NBAGR. It is high
Durbuk 145 27 28.0 10 +++++ probability of extinction of Ladakhi donkey
in Leh district in upcoming 60-70 years.
Total 5226 2628 11.5 67 ++++
(Contributed by Dr. S K Niranjan)
Genomic diversity and association study in indigenous cattle for adaptation
Whole genome SNP diversity analysis was performed
for four indigenous cattle breeds-Kankrej, Sir, Ladkhi,
and Hallikar representing the different agro-climatic
conditions utilizing various bioinformatics software
packages. SNPs were found to be most polymorphic
(85.33%) in Siri and least in Hallikar (57.33%). Maximum
heterozygosity was observed in Ladakhi (Ho= 0.40) and Siri
(Ho= 0.37) followed by Kankrej (Ho = 0.336) and lowest in
Hallikar (Ho = 0.330). Minor allele frequency (MAF) of hilly
Siri and Ladakhi cattle were significantly higher (p<0.05)
than those of Kankrej (0.24) and Hallikar (0.23), thus, Siri
having a good potential for their genetic development.
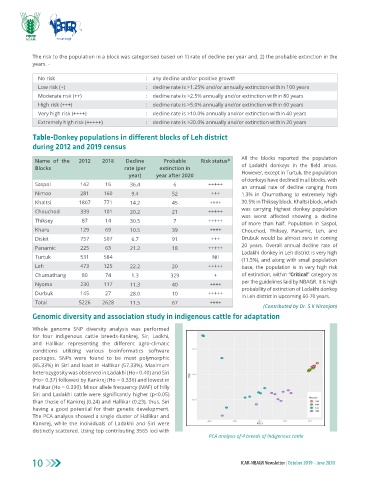
The PCA analysis showed a single cluster of Hallikar and
Kankrej, while the individuals of Ladakhi and Siri were
distinctly scattered. Using top contributing 3565 loci with
PCA analysis of 4 breeds of Indigenous cattle
10 ICAR-NBAGR Newsletter | October 2019 - June 2020