Page 11 - NBAGR NEWSLETTER VOL 16 No 1 (OCT 2019 - JUNE 2020)
P. 11
the largest F values (p-value<0.01), models having q-value of G-score less than 0.05 were considered as significant
ST
Bayesian algorithm-based STRUCTURE models. These significant models comprised of 30,350 unique SNPs. Variant effect
showed that individuals could be prediction (VEP) results revealed that 4,436 genes might have been influenced by
separated into two different groups. environmental variables.
The association study of the 46 samples (Contributed by Dr. Sanjeev Singh)
of 4 different cattle breeds (Hallikar,
Kankerej, Siri and Ladakhi) genotyped by Creation of DNA Chips for indigenous cattle and buffalo
bovine HD chip for the local adaptation
was carried out by R Sambada software Two DNA Chips (cattle and buffalo) specific to native breeds were developed for
with 5,74,382 SNPs which passed the genomic selection for increasing productivity. A total of 1676 animals belonging to
quality filter. A total of 11 environmental diverse breeds were sequenced at coverage of 10X yielding 60 GB of data per animal.
variables were finally taken in genome 196 SRS parentage markers were also tiled cattle HD DNA clip. The chip thus created
environment association study out of has been tested on 192 animals and has given a 99.8% call rate. The chip designed
which one variable was principal axis-1 has more than 585000 markers and is thus a high-density chip. The average distance
of the PCA of ancestry coefficients of the between the two markers is 4.57 Kb. Similarly, a high-density chip of Bubalus bubalis
individual’s samples. Other remaining has also been designed and is being analyzed for its performance.
variables consisted of longitude, latitude, (Contributed by Dr. RK Vijh)
Bio1, Bio2, Bio3, Bio4a, Bio12, tmax7,
prec10 and prec12. A total of 1,12,780
CRP-Genomics-Animal Component
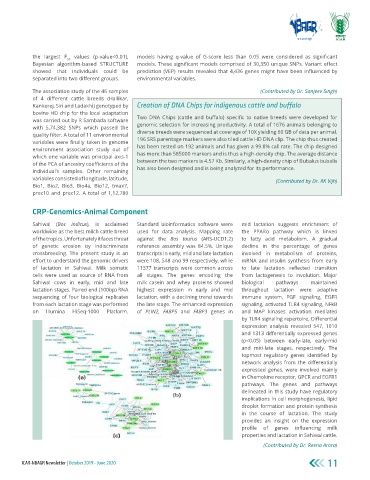
Sahiwal (Bos indicus), is acclaimed Standard bioinformatics software were mid lactation suggests enrichment of
worldwide as the best milch cattle breed used for data analysis. Mapping rate the PPARα pathway which is linked
of the tropics. Unfortunately it faces threat against the Bos taurus (ARS-UCD1.2) to fatty acid metabolism. A gradual
of genetic erosion by indiscriminate reference assembly was 84.5%. Unique decline in the percentage of genes
crossbreeding. The present study is an transcripts in early, mid and late lactation involved in metabolism of proteins,
effort to understand the genomic drivers were 108, 548 and 99 respectively, while mRNA and insulin synthesis from early
of lactation in Sahiwal. Milk somatic 11377 transcripts were common across to late lactation reflected transition
cells were used as source of RNA from all stages. The genes encoding the from lactogenesis to involution. Major
Sahiwal cows in early, mid and late milk casein and whey proteins showed biological pathways maintained
lactation stages. Paired end (100bp) RNA highest expression in early and mid throughout lactation were adaptive
sequencing of four biological replicates lactation, with a declining trend towards immune system, FGF signaling, EGFR
from each lactation stage was performed the late stage. The enhanced expression signaling, activated TLR4 signaling, NFkB
on Illumina HiSeq-1000 Platform. of PLIN2, FABP5 and FABP3 genes in and MAP kinases activation mediated
by TLR4 signaling repertoire. Differential
expression analysis revealed 547, 1010
and 1313 differentially expressed genes
(p<0.05) between early-late, early-mid
and mid-late stages, respectively. The
topmost regulatory genes identified by
network analysis from the differentially
expressed genes, were involved mainly
in Chemokine receptor, GPCR and EGFR1
pathways. The genes and pathways
delineated in this study have regulatory
implications in cell morphogenesis, lipid
droplet formation and protein synthesis
in the course of lactation. The study
provides an insight on the expression
profile of genes influencing milk
properties and lactation in Sahiwal cattle.
(Contributed by Dr. Reena Arora)
ICAR-NBAGR Newsletter | October 2019 - June 2020 11